|
chr17_-_32557916
|
64.362
|
NM_021319
|
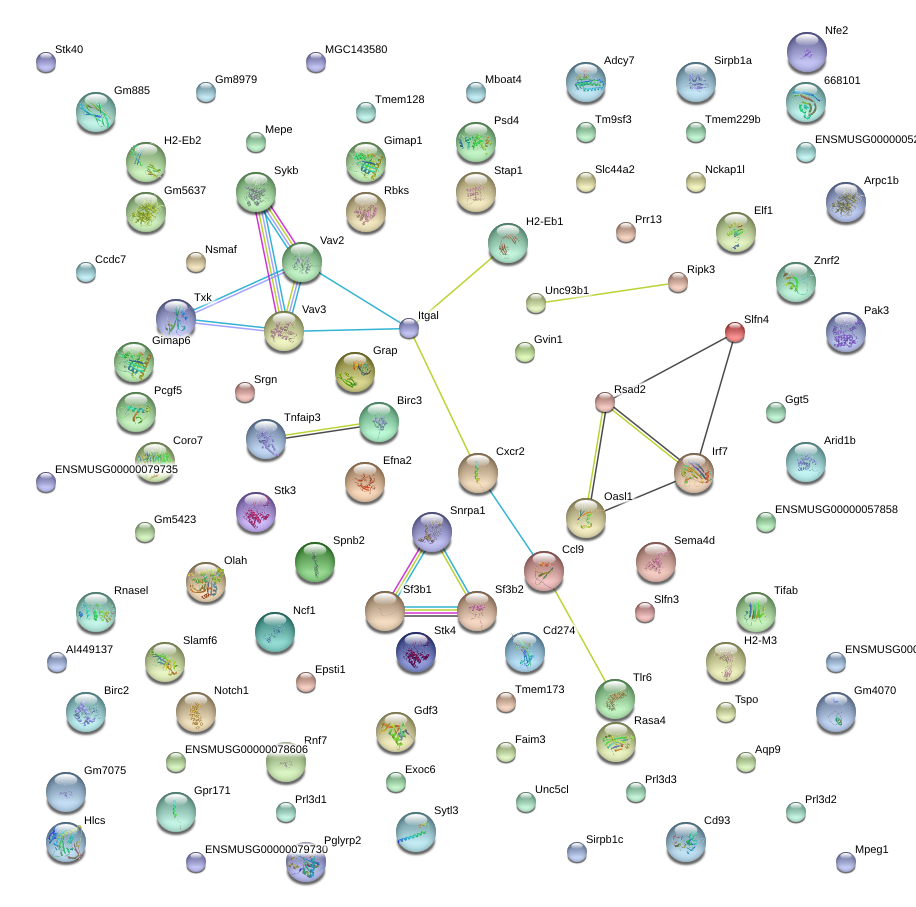
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr17_-_32557807
|
59.379
|
|
Pglyrp2
|
peptidoglycan recognition protein 2
|
|
chr15_+_103284200
|
37.257
|
NM_153505
|
Nckap1l
|
NCK associated protein 1 like
|
|
chr10_-_61960613
|
31.104
|
|
Srgn
|
serglycin
|
|
chr15_+_83393975
|
28.805
|
NM_009775
|
Tspo
|
translocator protein
|
|
chr2_-_27118325
|
28.310
|
|
Vav2
|
vav 2 oncogene
|
|
chr6_-_48658211
|
28.282
|
NM_153175
|
Gimap6
|
GTPase, IMAP family member 6
|
|
chr18_-_35900005
|
24.786
|
NM_028261
|
Tmem173
|
transmembrane protein 173
|
|
chr2_+_24242134
|
24.700
|
|
Psd4
|
pleckstrin and Sec7 domain containing 4
|
|
chr12_-_80084464
|
24.197
|
NM_001170401
|
Tmem229b
|
transmembrane protein 229B
|
|
chr14_-_56407622
|
23.882
|
NM_001164107
NM_001164108
NM_019955
|
Ripk3
|
receptor-interacting serine-threonine kinase 3
|
|
chr12_-_27141303
|
23.575
|
NM_021384
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr13_-_56280161
|
23.033
|
NM_001168615
NM_145976
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B
|
|
chr5_+_86500770
|
22.756
|
NM_019992
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr11_+_87569075
|
21.941
|
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene
|
|
chr5_-_73127743
|
21.538
|
NM_013698
|
Txk
|
TXK tyrosine kinase
|
|
chr17_+_34442837
|
21.292
|
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr7_+_134439804
|
20.348
|
NM_008400
|
Itgal
|
integrin alpha L
|
|
chr6_-_48658182
|
20.286
|
|
Gimap6
|
GTPase, IMAP family member 6
|
|
chr1_+_173847645
|
19.462
|
NM_030710
|
Slamf6
|
SLAM family member 6
|
|
chr11_+_82988687
|
19.353
|
NM_011410
|
Slfn4
|
schlafen 4
|
|
chr11_+_106612525
|
19.308
|
NM_001033435
|
Gm885
|
predicted gene 885
|
|
chr2_-_3283840
|
19.280
|
NM_145921
|
Olah
|
oleoyl-ACP hydrolase
|
|
chr19_-_41289262
|
18.952
|
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chr5_-_65351226
|
18.458
|
NM_011604
|
Tlr6
|
toll-like receptor 6
|
|
chr1_-_55044032
|
18.380
|
|
Sf3b1
|
splicing factor 3b, subunit 1
|
|
chr5_+_115373246
|
17.877
|
NM_145209
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr15_-_103085703
|
17.790
|
NM_008685
|
Nfe2
|
nuclear factor, erythroid derived 2
|
|
chr17_+_37407160
|
17.607
|
NM_013819
|
H2-M3
|
histocompatibility 2, M region locus 3
|
|
chr8_-_124948837
|
17.325
|
|
|
|
|
chr12_-_27141277
|
17.272
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr2_-_148263617
|
17.180
|
|
Cd93
|
CD93 antigen
|
|
chr3_-_58905678
|
17.040
|
NM_173398
|
Gpr171
|
G protein-coupled receptor 171
|
|
chr17_+_34442808
|
16.888
|
NM_010382
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr14_+_78304045
|
16.825
|
NM_029495
NM_178825
|
Epsti1
|
epithelial stromal interaction 1 (breast)
|
|
chr19_+_3935169
|
16.510
|
NM_001161428
NM_019449
|
Unc93b1
|
unc-93 homolog B1 (C. elegans)
|
|
chr6_-_122560002
|
16.225
|
|
Gdf3
|
growth differentiation factor 3
|
|
chr19_+_36453556
|
16.209
|
NM_029508
|
Pcgf5
|
polycomb group ring finger 5
|
|
chr17_+_6910178
|
16.145
|
NM_183368
|
Sytl3
|
synaptotagmin-like 3
|
|
chr17_+_48594225
|
16.103
|
NM_152823
|
Unc5cl
|
unc-5 homolog C (C. elegans)-like
|
|
chr6_-_122560022
|
15.907
|
NM_008108
|
Gdf3
|
growth differentiation factor 3
|
|
chr11_-_83388330
|
15.498
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr16_-_4679664
|
15.348
|
NM_030205
|
Coro7
|
coronin 7
|
|
chr4_+_125806078
|
15.300
|
|
Stk40
|
serine/threonine kinase 40
|
|
chr11_+_58680717
|
15.262
|
NM_172403
|
2810021J22Rik
|
RIKEN cDNA 2810021J22 gene
|
|
chr1_+_155596555
|
14.797
|
NM_011882
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent)
|
|
chr6_+_54839147
|
14.772
|
|
Znrf2
|
zinc and ring finger 2
|
|
chr7_-_148452299
|
14.625
|
NM_016850
|
Irf7
|
interferon regulatory factor 7
|
|
chr8_+_90812964
|
14.555
|
NM_001037723
|
Adcy7
|
adenylate cyclase 7
|
|
chr2_-_26314921
|
14.187
|
|
Notch1
|
Notch gene homolog 1 (Drosophila)
|
|
chr14_+_79915429
|
14.017
|
|
Elf1
|
E74-like factor 1
|
|
chr6_+_48689045
|
13.707
|
NM_008376
NM_175860
|
Gimap1
|
GTPase, IMAP family member 1
|
|
chr19_+_29441927
|
13.428
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr10_-_18731766
|
13.365
|
NM_001166402
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr19_+_12535536
|
13.265
|
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr17_+_5343877
|
13.164
|
|
Arid1b
|
AT rich interactive domain 1B (SWI-like)
|
|
chr10_+_75052084
|
13.119
|
NM_011820
|
Ggt5
|
gamma-glutamyltransferase 5
|
|
chr5_-_134705423
|
12.922
|
NM_010876
|
Ncf1
|
neutrophil cytosolic factor 1
|
|
chr7_+_73205419
|
12.736
|
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr9_+_21142267
|
12.695
|
NM_152808
|
Slc44a2
|
solute carrier family 44, member 2
|
|
chr5_+_145888588
|
12.551
|
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B
|
|
chr15_+_102289600
|
12.305
|
NM_001170911
NM_025385
|
Prr13
|
proline rich 13
|
|
chr2_+_163899842
|
12.239
|
NM_021420
|
Stk4
|
serine/threonine kinase 4
|
|
chr1_+_132762248
|
12.223
|
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr3_+_109376824
|
12.078
|
NM_146139
|
Vav3
|
vav 3 oncogene
|
|
chr8_-_131589372
|
12.023
|
NM_001197041
NM_029061
|
Ccdc7
|
coiled-coil domain containing 7
|
|
chr7_-_113358823
|
11.892
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr7_-_113102453
|
11.892
|
NM_001039160
NM_029000
|
Gvin1
|
GTPase, very large interferon inducible 1
|
|
chr19_+_12535268
|
11.871
|
NM_010821
|
Mpeg1
|
macrophage expressed gene 1
|
|
chr9_-_96378951
|
11.640
|
NM_011279
|
Rnf7
|
ring finger protein 7
|
|
chr17_+_35189798
|
11.602
|
|
|
|
|
chr5_+_104754347
|
11.589
|
NM_053172
|
Mepe
|
matrix extracellular phosphoglycoprotein with ASARM motif (bone)
|
|
chr8_+_77972592
|
11.544
|
NM_001164235
NM_027944
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene
|
|
chr13_-_51829951
|
11.512
|
|
Sema4d
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr5_+_115373272
|
11.294
|
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr5_+_136559785
|
11.240
|
NM_001039103
NM_133914
|
Rasa4
|
RAS p21 protein activator 4
|
|
chr10_+_127936302
|
11.225
|
|
A430046D13Rik
|
Riken cDNA A430046D13 gene
|
|
chr6_+_67966702
|
11.093
|
|
|
|
|
chr9_-_7872896
|
11.087
|
NM_007464
|
Birc3
|
baculoviral IAP repeat-containing 3
|
|
chr19_-_60302398
|
10.935
|
|
D19Ertd737e
|
DNA segment, Chr 19, ERATO Doi 737, expressed
|
|
chr13_+_52692221
|
10.926
|
NM_011518
|
Sykb
|
spleen tyrosine kinase
|
|
chr7_+_73205454
|
10.606
|
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A'
|
|
chr8_+_35178083
|
10.348
|
NM_001126314
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4
|
|
chr9_-_71011053
|
10.308
|
NM_022026
|
Aqp9
|
aquaporin 9
|
|
chr13_+_27248667
|
10.286
|
NM_172156
|
Prl3d3
|
prolactin family 3, subfamily d, member 3
|
|
chr16_-_94509462
|
9.940
|
NM_139145
|
Hlcs
|
holocarboxylase synthetase (biotin- [propriony-Coenzyme A-carboxylase (ATP-hydrolysing)] ligase)
|
|
chr13_-_97804287
|
9.914
|
NM_001080777
NM_025585
|
1700029F12Rik
|
RIKEN cDNA 1700029F12 gene
|
|
chr17_+_35403998
|
9.900
|
|
|
|
|
chr3_-_15426426
|
9.799
|
NM_001002898
|
Sirpb1a
|
signal-regulatory protein beta 1A
|
|
chr1_+_74200562
|
9.792
|
NM_009909
|
Cxcr2
|
chemokine (C-X-C motif) receptor 2
|
|
chr19_+_37624879
|
9.711
|
NM_175353
|
Exoc6
|
exocyst complex component 6
|
|
chr10_+_126504125
|
9.641
|
|
|
|
|
chr11_+_61466766
|
9.614
|
|
Grap
|
GRB2-related adaptor protein
|
|
chr4_-_6381171
|
9.609
|
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr5_+_38651466
|
9.605
|
NM_025480
|
Tmem128
|
transmembrane protein 128
|
|
chr5_-_31950177
|
9.579
|
|
Rbks
|
ribokinase
|
|
chr8_+_131591033
|
9.574
|
NM_001168369
NM_029292
|
1700008F21Rik
|
RIKEN cDNA 1700008F21 gene
|
|
chr1_-_146024404
|
9.523
|
NM_153171
|
Rgs13
|
regulator of G-protein signaling 13
|
|
chr9_-_32348863
|
9.481
|
NM_008026
|
Fli1
|
Friend leukemia integration 1
|
|
chr7_-_48755259
|
9.424
|
NM_001033530
|
AW146154
|
expressed sequence AW146154
|
|
chr9_+_114640266
|
9.413
|
NM_026036
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr11_-_46202942
|
9.366
|
NM_010583
|
Itk
|
IL2-inducible T-cell kinase
|
|
chr11_-_98190774
|
9.234
|
NM_010895
|
Neurod2
|
neurogenic differentiation 2
|
|
chr3_-_15748418
|
9.126
|
NM_001173459
|
LOC100038947
Sirpb1a
|
signal-regulatory protein beta 1-like
signal-regulatory protein beta 1A
|
|
chr10_-_111434265
|
9.096
|
NM_028608
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr17_-_34133397
|
9.036
|
|
H2-K1
|
histocompatibility 2, K1, K region
|
|
chr15_-_9459612
|
8.995
|
NM_008372
|
Il7r
|
interleukin 7 receptor
|
|
chr3_-_144347983
|
8.992
|
|
|
|
|
chr11_+_61466797
|
8.991
|
NM_027817
|
Grap
|
GRB2-related adaptor protein
|
|
chr5_+_114580397
|
8.988
|
NM_001040691
|
Ung
|
uracil DNA glycosylase
|
|
chr5_+_106039499
|
8.936
|
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C
|
|
chr10_-_111434231
|
8.926
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr19_-_44210311
|
8.867
|
NM_001081077
|
Cwf19l1
|
CWF19-like 1, cell cycle control (S. pombe)
|
|
chr1_+_133535033
|
8.848
|
|
Ctse
|
cathepsin E
|
|
chr6_-_129225348
|
8.690
|
NM_001033122
|
Cd69
|
CD69 antigen
|
|
chr19_-_27463532
|
8.675
|
|
|
|
|
chr4_+_128688217
|
8.605
|
|
|
|
|
chr7_-_34977928
|
8.506
|
|
Pdcd2l
|
programmed cell death 2-like
|
|
chr19_-_30623890
|
8.505
|
NM_010051
|
Dkk1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr19_+_12873115
|
8.494
|
|
Lpxn
|
leupaxin
|
|
chr8_+_23348971
|
8.453
|
NM_019576
|
Thsd1
|
thrombospondin, type I, domain 1
|
|
chr2_+_153956082
|
8.345
|
NM_028528
|
1700058C13Rik
|
RIKEN cDNA 1700058C13 gene
|
|
chr6_+_100783598
|
8.336
|
NM_182939
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr17_+_87316687
|
8.313
|
NM_030104
|
1700090G07Rik
|
RIKEN cDNA 1700090G07 gene
|
|
chr6_-_129572590
|
8.308
|
NM_033078
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr14_+_27813547
|
8.302
|
NM_010420
|
Hesx1
|
homeobox gene expressed in ES cells
|
|
chr19_+_12873063
|
8.297
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr9_+_123715592
|
8.261
|
NM_030712
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6
|
|
chr17_+_33102747
|
8.208
|
NM_153063
|
Zfp472
|
zinc finger protein 472
|
|
chr5_+_115346942
|
8.205
|
NM_011854
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr4_-_149072755
|
8.184
|
NM_001164049
NM_001164050
NM_008840
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr3_-_106592919
|
8.144
|
NM_007651
|
Cd53
|
CD53 antigen
|
|
chr1_+_109407998
|
8.143
|
NM_001174170
NM_011111
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2
|
|
chr9_+_38525852
|
8.121
|
NM_001145957
NM_172767
|
Vwa5a
|
von Willebrand factor A domain containing 5A
|
|
chrX_-_162948399
|
8.088
|
NM_001007577
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr13_-_64230610
|
8.069
|
NM_001001321
|
Slc35d2
|
solute carrier family 35, member D2
|
|
chrX_-_7544940
|
8.066
|
NM_008089
|
Gata1
|
GATA binding protein 1
|
|
chrX_-_154006916
|
8.062
|
NM_001098723
|
Yy2
Mbtps2
|
Yy2 transcription factor
membrane-bound transcription factor peptidase, site 2
|
|
chr9_-_62644026
|
8.009
|
|
Fem1b
|
feminization 1 homolog b (C. elegans)
|
|
chr7_-_25873560
|
7.965
|
|
|
|
|
chr2_-_164182009
|
7.964
|
|
Slpi
|
secretory leukocyte peptidase inhibitor
|
|
chr4_-_149075697
|
7.867
|
NM_001029837
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr2_+_32583221
|
7.839
|
|
Sh2d3c
|
SH2 domain containing 3C
|
|
chr11_+_53164267
|
7.794
|
NM_033565
|
Aff4
|
AF4/FMR2 family, member 4
|
|
chr12_+_75028976
|
7.783
|
|
Hif1a
|
hypoxia inducible factor 1, alpha subunit
|
|
chr7_-_16807341
|
7.781
|
NM_027883
|
Dhx34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34
|
|
chr16_+_44811858
|
7.657
|
|
Cd200r4
|
CD200 receptor 4
|
|
chr6_-_118089060
|
7.603
|
NM_030165
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2
|
|
chr2_-_118554094
|
7.581
|
NM_177568
|
Plcb2
|
phospholipase C, beta 2
|
|
chr16_+_59471600
|
7.548
|
NM_025910
|
Mina
|
myc induced nuclear antigen
|
|
chr11_-_99855307
|
7.464
|
NM_001099774
|
Krtap17-1
|
keratin associated protein 17-1
|
|
chr1_+_52687548
|
7.396
|
NM_001142647
|
Tmem194b
|
transmembrane protein 194B
|
|
chr9_+_43552516
|
7.394
|
NM_021424
|
Pvrl1
|
poliovirus receptor-related 1
|
|
chr7_+_134439888
|
7.345
|
|
Itgal
|
integrin alpha L
|
|
chrX_+_156411885
|
7.340
|
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr17_+_37006518
|
7.326
|
NM_011280
|
Trim10
|
tripartite motif-containing 10
|
|
chr2_-_91019477
|
7.312
|
NM_001177720
|
Madd
|
MAP-kinase activating death domain
|
|
chr13_+_95335701
|
7.274
|
|
|
|
|
chr1_-_80661844
|
7.256
|
|
Dock10
|
dedicator of cytokinesis 10
|
|
chr12_+_72149332
|
7.252
|
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr9_-_88626434
|
7.197
|
|
Bcl2a1d
|
B-cell leukemia/lymphoma 2 related protein A1d
|
|
chr16_+_91465283
|
7.173
|
|
A930006K02Rik
|
RIKEN cDNA A930006K02 gene
|
|
chr2_-_60589953
|
7.172
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_-_65323937
|
7.172
|
NM_030682
|
Tlr1
|
toll-like receptor 1
|
|
chr13_-_53472732
|
7.166
|
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1
|
|
chr6_+_83293025
|
7.145
|
|
Mobkl1b
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr14_+_21520038
|
7.090
|
|
|
|
|
chr1_-_173579128
|
7.044
|
|
Slamf7
|
SLAM family member 7
|
|
chr6_-_115685120
|
7.044
|
NM_001168258
|
Tmem40
|
transmembrane protein 40
|
|
chr9_-_14185399
|
6.985
|
|
Endod1
|
endonuclease domain containing 1
|
|
chr2_-_154196611
|
6.970
|
|
Cdk5rap1
|
CDK5 regulatory subunit associated protein 1
|
|
chr1_+_161974815
|
6.968
|
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene
|
|
chrX_+_70437517
|
6.945
|
NM_001081643
|
Xlr3b
|
X-linked lymphocyte-regulated 3B
|
|
chr17_+_22498419
|
6.944
|
NM_145484
|
Zfp758
|
zinc finger protein 758
|
|
chr3_+_95466898
|
6.918
|
|
|
|
|
chr18_-_32719516
|
6.890
|
NM_001048207
|
Gypc
|
glycophorin C
|
|
chr10_+_96079634
|
6.864
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr1_-_173579057
|
6.863
|
|
Slamf7
|
SLAM family member 7
|
|
chr6_+_83065466
|
6.833
|
NM_020619
|
Mogs
|
mannosyl-oligosaccharide glucosidase
|
|
chr1_+_132762353
|
6.830
|
NM_026976
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr6_+_53523365
|
6.816
|
NM_172728
|
Creb5
|
cAMP responsive element binding protein 5
|
|
chr5_+_137170046
|
6.808
|
NM_001085387
|
Myl10
|
myosin, light chain 10, regulatory
|
|
chr17_-_50329593
|
6.720
|
NM_181397
|
Rftn1
|
raftlin lipid raft linker 1
|
|
chr17_-_51878720
|
6.709
|
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr2_+_132516018
|
6.697
|
NM_028637
|
1110034G24Rik
|
RIKEN cDNA 1110034G24 gene
|
|
chr12_+_82049290
|
6.691
|
|
Srsf5
|
serine/arginine-rich splicing factor 5
|
|
chr9_-_14185685
|
6.684
|
NM_028013
|
Endod1
|
endonuclease domain containing 1
|
|
chr1_-_173537404
|
6.682
|
NM_008534
|
Ly9
|
lymphocyte antigen 9
|
|
chr13_-_53472719
|
6.632
|
NM_009269
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1
|
|
chr19_-_41289045
|
6.593
|
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chr9_+_7792286
|
6.566
|
|
Tmem123
|
transmembrane protein 123
|
|
chr17_+_35404109
|
6.566
|
|
|
|
|
chr4_+_134483092
|
6.555
|
|
|
|
|
chr9_-_71743854
|
6.532
|
|
Tcf12
|
transcription factor 12
|
|
chrX_+_104284572
|
6.517
|
NM_172435
|
P2ry10
|
purinergic receptor P2Y, G-protein coupled 10
|
|
chr9_-_21767872
|
6.493
|
NM_010149
|
Epor
|
erythropoietin receptor
|
|
chr4_-_43666412
|
6.477
|
NM_001007463
|
Spag8
|
sperm associated antigen 8
|
|
chr17_+_28667805
|
6.474
|
NM_026290
|
4930511I11Rik
|
RIKEN cDNA 4930511I11 gene
|
|
chr5_+_30969259
|
6.423
|
NM_007681
|
Cenpa
|
centromere protein A
|
|
chr6_-_52190771
|
6.403
|
NM_001122950
|
Hoxa10
|
homeobox A10
|